Network Medicine: a brief introduction
Exploratory: Network Medicine
Genes disease associations have been identified by genome-wide association studies (GWAS). Unluckily, our knowledge of the mechanisms underlying these associations that are responsible for the diseases remains largely undefined. There is increasing evidence that a set of proteins associated with a disease do not work in an isolated way, but they interact with each other to form a distinct networkmodule representing perturbed and dysfunctional pathways.
Network medicine is an emerging area of research dealing with molecular and genetic interactions, network biomarkers of disease, and therapeutic target discovery. In many applications, Network Medicine seeks to use cellular molecular pathways to explore the etiologies of human diseases. However, since many molecular pathways remain poorly defined, Network Medicine relies on inference or interaction networks, and then uses the resulting networks to explore drivers of disease.
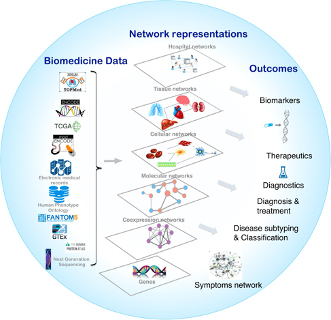
Figure[1] Overview of network medicine approach depicting various biomedical data types, along with network representations that simplify different components of multiple omics data from the genome, transcriptome, proteome, and metabolome as nodes that are connected by links (edges). Combining biomedical data with the appropriate network modeling approach allows derivation of disease associated information and outcomes like biomarkers, therapeutics targets, phenotype-specific genes and interactions, and disease subtypes.
Large-scale biomedical data generation offers a unique opportunity to assess the effect and impact of cellular heterogeneity and environmental perturbations on the observed phenotype. To understand a given phenotype, the functioning of a cell or the cellular organization, network medicine relies mostly on measurements of the abundance profiles inside the cells. The analysis of this abundance allows us to infer the interactions leading to the observed phenotype. Different biological networks capture the complex interactions between genes, proteins, RNA molecules, metabolites and genetic variants in the cells of organisms.
Network Medicine focuses on disease biology, including understanding disease etiology, identifying potential biomarkers, and designing therapeutic interventions, including drug targets, dosage, and synergism discovery. Indeed, the ultimate goal is to develop a global understanding of the propagation of perturbations in the system by identifying the pathways, subtypes of disease states, and key components in the networks that can be targeted in clinical interventions[1].
Constructing appropriate network models is a challenging problem that heavily depends on the study design, the phenotype under study, the molecular entities measured, and the type and size of the data. The field of network medicine is largely discovery — rather than hypothesis — driven, uncovering previously unknown relationships and leading to the identification of new biomarkers and new insights into the molecular mechanisms of the given phenotype, such as tissue specificity or disease context. The statistical rigor of network predictions comes from the study design and the size of the datasets.
A variety of different methods has been used to infer relevant cellular molecular networks. Protein–protein interaction networks[2], have been used to identify interconnected subsets of interacting proteins related to specific diseases, known as disease network modules[3]. Methods such as weighted gene coexpression network analysis[4] use pairwise correlation in gene expression levels to identify modules of similarly expressed genes in distinct phenotypic states. Still, other methods integrate multiple different data types to infer regulatory interactions between transcription factors and their targets[5]. These modeling approaches based on Big Data can be thought of as “top-down” efforts to identify genes of interest agnostically in disease-related networks.
In conclusion improvements in disease diagnosis will have the most immediate impact on clinical medicine. However, substantial advancements in the analysis, interpretation, and validation of Network Medicine approaches will be required to turn this potential to transform medical care into reality.
This article is inspired by Sonawane, Abhijeet R., et al. "Network medicine in the age of biomedical big data." Frontiers in Genetics 10 (2019): 294.
Written by: Michele Gentili
Revised by: Luca Pappalardo
[1] Sonawane, Abhijeet R., et al. "Network medicine in the age of biomedical big data." Frontiers in Genetics 10 (2019): 294.
[2] Goh, K. I., Cusick, M. E., Valle, D., Childs, B., Vidal, M., and Barabasi, A. L. (2007). The human disease network. Proc. Natl. Acad. Sci. U.S.A. 104, 8685–8690. doi: 10.1073/pnas.0701361104
[3] Gentili, et al. "Biological Random Walks: Integrating heterogeneous data in disease gene prioritization." 2019 (CIBCB). IEEE, 2019.
[4] Langfelder, P., and Horvath, S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559. doi: 10.1186/1471-2105-9-559
[5] Glass, K., Huttenhower, C., Quackenbush, J., and Yuan, G. C. (2013). Passing messages between biological networks to refine predicted interactions. PLoS One 8:e64832. doi: 10.1371/journal.pone.0064832
[1] Sonawane, Abhijeet R., et al. "Network medicine in the age of biomedical big data." Frontiers in Genetics 10 (2019): 294.

